Extending The Grammar of Graphics for Biological Data Visualization

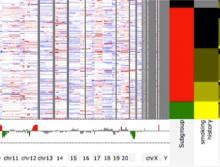
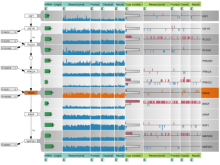
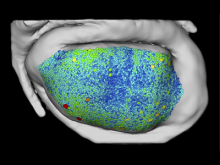
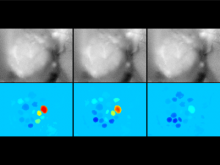
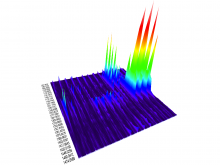
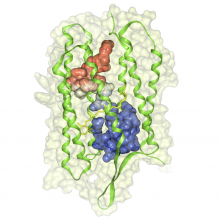
This work extends the grammar of graphics into the domain of genomic data visualization, focusing on next generation sequencing data. It develops object oriented design to utilize charting taxonomies with a goal to improve consistency in plots of biological data. The work builds on the grammar of graphics in ggplot2 in R, and makes use of the extensive statistical functionality and data processing in Bioconductor. The new R package is called "ggbio". This work aims to shed light on biology-specific visualization and the design of graphic with biological data analysis and modeling.
BioVis 2012 Information