Visualization and Exploration of 3D Toponome Data
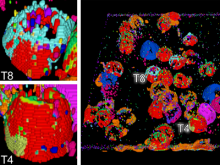
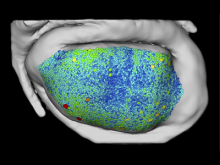
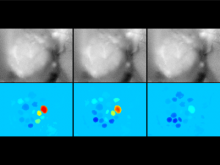
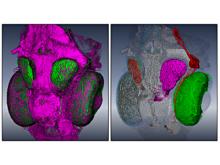
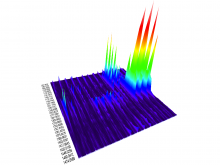
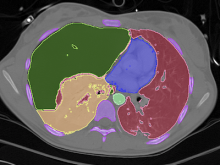
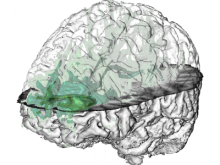
In toponomics, the toponome, i.e. the location and topological distribution of proteins across a cell, is investigated. Toponome imaging results in a 3D volume per protein affinity reagent. After imaging, the data is binarized such that 1 encodes protein present and 0 encodes protein absent. We present a volume rendering approach for visualizing all unique binary protein patterns. A unique color is dynamically assigned to each pattern such that a sufficient perceptual difference between colors in the current view is guaranteed. We further present techniques for interacting with the view. We demonstrate our approach for a cell sample containing lymphocytes.
BioVis 2012 Information